How to merge data in R using R merge, dplyr, or data.table
source link: https://www.infoworld.com/article/3454356/how-to-merge-data-in-r-using-r-merge-dplyr-or-datatable.html
Go to the source link to view the article. You can view the picture content, updated content and better typesetting reading experience. If the link is broken, please click the button below to view the snapshot at that time.
R has a number of quick, elegant ways to join data frames by a common column. I’d like to show you three of them:
- base R’s
merge()function dplyr’s join family of functionsdata.table’s bracket syntax
Get and import the data
For this example I’ll use one of my favorite demo data sets—flight delay times from the U.S. Bureau of Transportation Statistics. If you want to follow along, head to http://bit.ly/USFlightDelays and download data for the time frame of your choice with the columns Flight Date, Reporting_Airline, Origin, Destination, and DepartureDelayMinutes. Also get the lookup table for Reporting_Airline.
Or, you can download these two data sets—plus my R code in a single file and a PowerPoint explaining different types of data merges—here:
To read in the file with base R, I’d first unzip the flight delay file and then import both flight delay data and the code lookup file with read.csv(). If you’re running the code, the delay file you downloaded will likely have a different name than in the code below. Also, note the lookup file’s unusual .csv_ extension.
unzip("673598238_T_ONTIME_REPORTING.zip")
mydf <- read.csv("673598238_T_ONTIME_REPORTING.csv",
sep = ",", quote="\"")
mylookup <- read.csv("L_UNIQUE_CARRIERS.csv_",
quote="\"", sep = "," )Next, I’ll take a peek at both files with head():
head(mydf)
FL_DATE OP_UNIQUE_CARRIER ORIGIN DEST DEP_DELAY_NEW X
1 2019-08-01 DL ATL DFW 31 NA
2 2019-08-01 DL DFW ATL 0 NA
3 2019-08-01 DL IAH ATL 40 NA
4 2019-08-01 DL PDX SLC 0 NA
5 2019-08-01 DL SLC PDX 0 NA
6 2019-08-01 DL DTW ATL 10 NA
head(mylookup)
Code Description
1 02Q Titan Airways
2 04Q Tradewind Aviation
3 05Q Comlux Aviation, AG
4 06Q Master Top Linhas Aereas Ltd.
5 07Q Flair Airlines Ltd.
6 09Q Swift Air, LLC d/b/a Eastern Air Lines d/b/a EasternMerges with base R
The mydf delay data frame only has airline information by code. I’d like to add a column with the airline names from mylookup. One base R way to do this is with the merge() function, using the basic syntax merge(df1, df2). The order of data frame 1 and data frame 2 doesn't matter, but whichever one is first is considered x and the second one is y.
If the columns you want to join by don’t have the same name, you need to tell merge which columns you want to join by: by.x for the x data frame column name, and by.y for the y one, such as merge(df1, df2, by.x = "df1ColName", by.y = "df2ColName").
You can also tell merge whether you want all rows, including ones without a match, or just rows that match, with the arguments all.x and all.y. In this case, I’d like all the rows from the delay data; if there’s no airline code in the lookup table, I still want the information. But I don’t need rows from the lookup table that aren’t in the delay data (there are some codes for old airlines that don’t fly anymore in there). So, all.x equals TRUE but all.y equals FALSE. Here's the code:
joined_df <- merge(mydf, mylookup, by.x = "OP_UNIQUE_CARRIER",
by.y = "Code", all.x = TRUE, all.y = FALSE)The new joined data frame includes a column called Description with the name of the airline based on the carrier code:
head(joined_df)
OP_UNIQUE_CARRIER FL_DATE ORIGIN DEST DEP_DELAY_NEW X Description
1 9E 2019-08-12 JFK SYR 0 NA Endeavor Air Inc.
2 9E 2019-08-12 TYS DTW 0 NA Endeavor Air Inc.
3 9E 2019-08-12 ORF LGA 0 NA Endeavor Air Inc.
4 9E 2019-08-13 IAH MSP 6 NA Endeavor Air Inc.
5 9E 2019-08-12 DTW JFK 58 NA Endeavor Air Inc.
6 9E 2019-08-12 SYR JFK 0 NA Endeavor Air Inc.Joins with dplyr
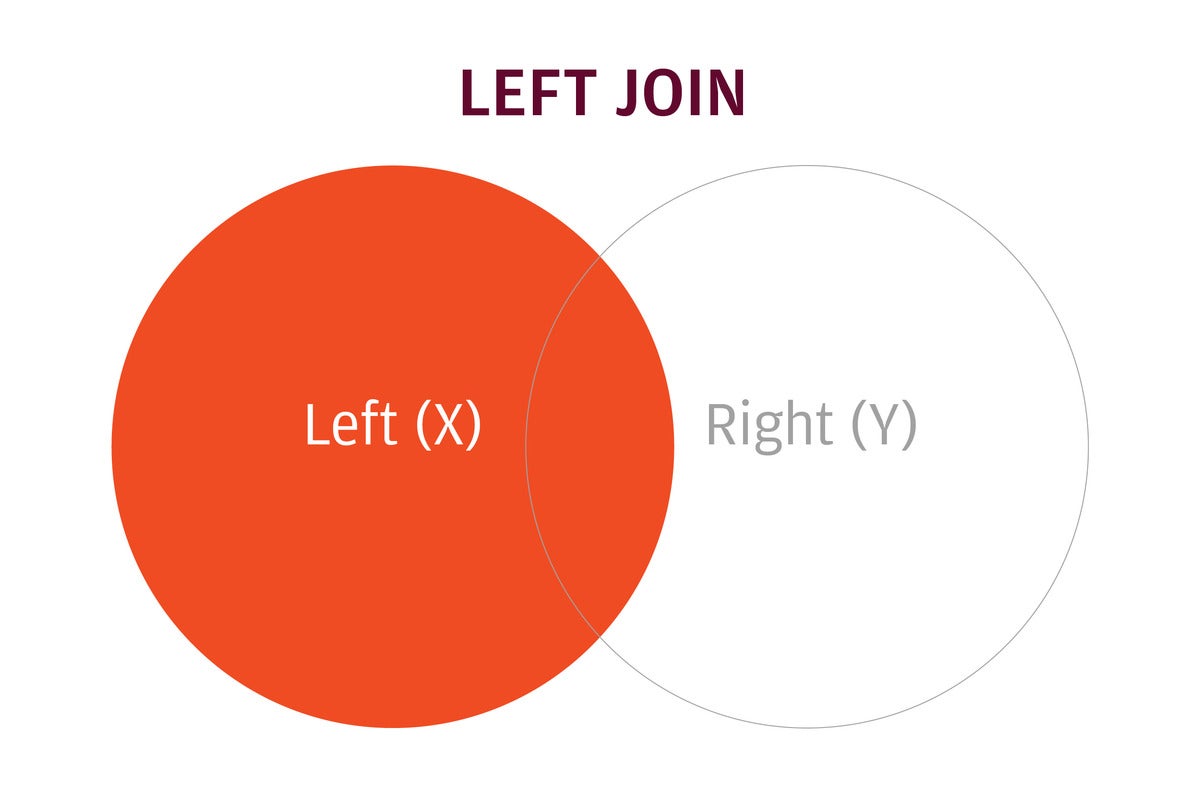
The dplyr package uses SQL database syntax for its join functions. A left join means: Include everything on the left (what was the x data frame in merge()) and all rows that match from the right (y) data frame. If the join columns have the same name, all you need is left_join(x, y). If they don’t have the same name, you need a by argument, such as left_join(x, y, by = c("df1ColName" = "df2ColName")).
Note the syntax for by: It’s a named vector, with both the left and right column names in quotation marks.
Update: Starting with dplyr version 1.1.0 (on CRAN as of January 29, 2023), dplyr joins have an additional by syntax using join_by():
left_join(x, y, by = join_by(df1ColName == df2ColName))The new join_by() helper function uses unquoted column names and the == boolean operator, which package authors say makes more sense in an R context than c("col1" = "col2"), since = is meant for assigning a value to a variable, not testing for equality.
IDG
A left join keeps all rows in the left data frame and only matching rows from the right data frame.
The code to import and merge both data sets using left_join() is below. It starts by loading the dplyr and readr packages, and then reads in the two files with read_csv(). When using read_csv(), I don’t need to unzip the file first.
library(dplyr)
library(readr)
mytibble <- read_csv("673598238_T_ONTIME_REPORTING.zip")
mylookup_tibble <- read_csv("L_UNIQUE_CARRIERS.csv_")joined_tibble <- left_join(mytibble, mylookup_tibble,
by = join_by(OP_UNIQUE_CARRIER == Code))
Note that dplyr's older by syntax without join_by() still works
joined_tibble <- left_join(mytibble, mylookup_tibble,
by = c("OP_UNIQUE_CARRIER" = "Code"))
read_csv() creates tibbles, which are a type of data frame with some extra features. left_join() merges the two. Take a look at the syntax: In this case, order matters. left_join() means include all rows on the left, or first, data set, but only rows that match from the second one. And, because I need to join by two differently named columns, I included a by argument.
The new join syntax in the development-only version of dplyr would be:
joined_tibble2 <- left_join(mytibble, mylookup_tibble,
by = join_by(OP_UNIQUE_CARRIER == Code))Since most people likely have the CRAN version, however, I will use dplyr's original named-vector syntax in the rest of this article, until join_by() becomes part of the CRAN version.
We can look at the structure of the result with dplyr’s glimpse() function, which is another way to see the top few items of a data frame:
glimpse(joined_tibble)
Observations: 658,461
Variables: 7
$ FL_DATE <date> 2019-08-01, 2019-08-01, 2019-08-01, 2019-08-01, 2019-08-01…
$ OP_UNIQUE_CARRIER <chr> "DL", "DL", "DL", "DL", "DL", "DL", "DL", "DL", "DL", "DL",…
$ ORIGIN <chr> "ATL", "DFW", "IAH", "PDX", "SLC", "DTW", "ATL", "MSP", "JF…
$ DEST <chr> "DFW", "ATL", "ATL", "SLC", "PDX", "ATL", "DTW", "JFK", "MS…
$ DEP_DELAY_NEW <dbl> 31, 0, 40, 0, 0, 10, 0, 22, 0, 0, 0, 17, 5, 2, 0, 0, 8, 0, …
$ X6 <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ Description <chr> "Delta Air Lines Inc.", "Delta Air Lines Inc.", "Delta Air …This joined data set now has a new column with the name of the airline. If you run a version of this code yourself, you’ll probably notice that dplyr is way faster than base R.
Next, let’s look at a super-fast way to do joins.
Recommend
-
 25
25
1. dplyr简介 dplyr是R语言的数据分析包,类似于python中的pandas,能对dataframe类型的数据做很方便的...
-
 6
6
An orderly approach to regression models, ideally with dplyr advertisements Reading the documentation for do() in dplyr, I've bee...
-
 12
12
How to merge data in Python using Pandas merge Use the popular Pandas library for data manipulation and analysis to read data from two files and join them into a single dataset.
-
 8
8
Using NodeRed to publish data to Azure Table Storage
-
 2
2
Raphael WALTER August 17, 2023 8 minute read...
-
 2
2
Merge two images of data in a group with Group By advertisements I have two dataframes and need to merge them based on a date, but the merge should...
-
 2
2
Mustafa Uzumcu February 8, 2022 5 minute read ...
-
 2
2
Data Migration Project: Portal Merge March 3, 2022 by Giedrius Ramašauskas At Vinted, our long-term strategy...
-
 32
32
pandas merge(): Combining Data on Common Columns or Indices The first technique that you’ll learn is merge(). You can use merge() anytime you want functionality similar to a database’s
-
 3
3
Combining Data in pandas With concat() and merge() The Series and DataFrame objects in pandas are powerful tools for exploring and analyzing data. Part of their power comes from a multifaceted approach to combin...
About Joyk
Aggregate valuable and interesting links.
Joyk means Joy of geeK