

Can we predict a pathway status using gene expression?
source link: https://www.neuraldesigner.com/blog/kras_mutation_inference_based_on_expression
Go to the source link to view the article. You can view the picture content, updated content and better typesetting reading experience. If the link is broken, please click the button below to view the snapshot at that time.

Can we predict a pathway status using gene expression?.
Can we predict a pathway status using gene expression?

Abnormal pathway activation may lead to diseases. In this context, if we assume that a mutation in a core gene leads to that status, can we predict it using expression data?.
The original data used in this example is stored in the GDC web, where the Cancer Genome Atlas (TCGA) Research Network has profiled and analyzed large numbers of human tumors to discover molecular aberrations at the DNA, RNA, protein and epigenetic levels.
We will be using Neural Designer free version to try to answer the previous question. You can download a free trial here.
Introduction
A pathway status describes a series of chemical reactions where cell molecules work together to control a cell function, such as cell division or death.
For example, cells receive a signal from their environment when a molecule joins a specific protein receptor on or in the cell. After the first molecule in the pathway receives a signal, it activates another molecule. This process of sequential activation is repeated along the signaling pathway until the last molecule is activated and the function is carried out.
However, the abnormal activation of pathways may lead to diseases such as cancer. In this example, we will be focused on the RTK/RAS/PI3K pathway:

This molecular genetic axis controls critical cellular functions and is commonly altered in various cancers. Perturbations across this pathway can lead to deficiencies in the cell cycle, survival, metabolism, motility, and genome stability, triggering cancer hallmark phenotypes.
Specifically, we will look at KRAS mutations. So if KRAS is mutated, we consider the pathway to be altered. At this point, we wonder: would we be able to predict whether a pathway is altered based on gene expression data alone?.
Materials and methods
Dataset
This dataset contains the analys of large numbers of human tumors to discover molecular aberrations at the DNA, RNA, protein and epigenetic levels made by the TCGA. This information is reported for 9074 patients and for 33 different cancer types. We will focused on the expression data obtained by RNA-seq and the mutational data.
We will download the data from Zenodo, where the raw files have been processed to tabular format. We will be using the files for the expressions (pancan_rnaseq_freeze.tsv.gz) and the file for the mutations (pancan_mutation_freeze.tsv.gz).

The final merged data has 9074 rows, corresponding to patients suffering from one of 33 cancer types that TCGA reports. Each patient also has expression data for 20501 genes, and lastly, a binary variable that indicates whether or not this patient has the pathway altered.
This variable was calculated considering KRAS mutations. Therefore, if this gene is mutated for a patient, we assume the pathway is altered.
Next, we are going to split our data into two subsets: one with genes inside the pathway and another one with the genes outside the pathway. This gene list is taken from Kegg.
We filter our data, and divide it into a 350 and a 20151 gene datasets, corresponding to genes inside or outside the pathway. Also, we will maintain samples where only KRAS is mutated and where neither of the three genes of the pathway is mutated. In the end, we have 7803 samples.
Once we have these two datasets, we will follow the Neural Designer workflow to generate two models for each dataset.
Model development
First, as we will be comparing two models, for robustness, we will use the same optimization algorithm, loss index and the same network structure for both.

We will be using a network with genes as input variables and a scaling layer with the same number of neurons as the number of inputs we have in each case. We will use ten neurons with the hyperbolic tangent as the activation function for the perceptron layer.
This number of neurons results from previous testing with the data to obtain better model regularization. Finally, our probabilistic layer with one neuron gives us the probability of the pathway being altered.
In the training strategy, we will use the Weighted squared error for the loss index because we have a varying group size for the predicted variable (703 vs. 7100). In addition, we will use the Quasi-Newton method for the optimization algorithm, as it is the standard for classification problems.
Finally, we will use feature selection on our network to reduce the input genes to a more manageable size.
Results
We have built two models for each of the subsets mentioned above. In this models we are using gene expression of a subset of genes to predict wether or not the target pathway is altered.
Model for genes inside the pathway
In this model we have the genes that are inside the pathway. With these genes we are able to predict wether or not the pathway is altered. The genes included are: AKT3, AREG, BCL2, CREB3L1, EFNA2, EFNA3, EFNA4, EPHA2, EREG, FGF18, FGF19, FGFR4, GHR, GNG7, IL2RG, IL7, INS, ITGA2, ITGB4, ITGB6, ITGA7, LAMA3, LAMB3, LAMC2, MAGI2, MYB, NGFR.
Next we will use a ROC curve to asses if our model performs adequately. The ROC curve is a standard method for testing a neural network using an independent subset of the original data in binary classification applications, it plots false positives rate (or 1 - specificity) on the X-axis, and true negatives rate (or sensitivity) on the Y-axis for different decision threshold values. The following figure shows the ROC curve for the genes inside the pathway model:

As we can see in the previous image we have a good ROC curve, meaning that we a have a working model. For this ROC curve, we obtain the following confusion matrix where we represent how the testing samples are classified with our model:
| τ=0.5 | Predicted positive | Predicted Negative |
|---|---|---|
| Real positive | 102 (6.53%) | 39 (2.5%) |
| Real negative | 153 (9.8%) | 1266 (81.15%) |
From this matrix we obtain the specificity and the sensitivity of the model: Sensitivity: 0.72, Specificity: 0.89. All this information implies that this model is working as intended and we can obtain good results with it.
Model for genes outside the pathway
In this model we have the genes that are outside the pathway. With these genes we are able to predict wether or not the pathway is altered. The genes included are: ALPP, AP3B2, BTNL8, BSPRY, C3orf54, C10orf81, C10orf95, C10orf108, C11orf53, C14orf50, C14orf72, C15orf48, C17orf73, C19orf22, C19orf33, C19orf45, C20orf118, C20orf151, CDIPT, MLKL, RAB11FIP1, SDCBP2, SLC44A4, SMPDL3B, SPATA12, SPRY2, SRPK1.
Next we will use a ROC curve to asses if our model performs adequately. The following figure shows the ROC curve for the genes inside the pathway model:
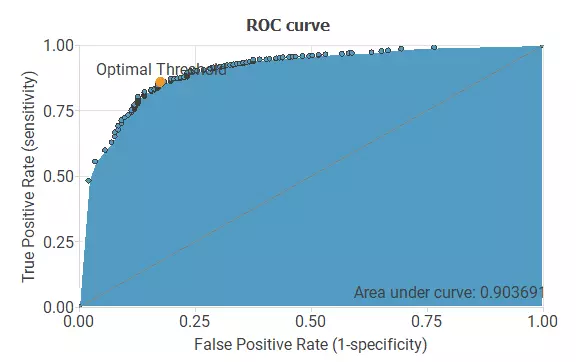
As we can see in the previous image we get a good ROC curve, meaning that we a have a working model. Also, we get a slightly better ROC curve than in the previous model.
And the confusion matrix for this model:
| τ=0.5 | Predicted positive | Predicted Negative |
|---|---|---|
| Real positive | 93 (5.9%) | 45 (2.88%) |
| Real negative | 135 (8.6%) | 1287 (82.5% |
Also, this model has a slightly worse sensitivity but a better specificity than the previous model: Sensitivity: 0.67, Specificity: 0.90. In the end, we have two working models, one with genes inside the case study pathway and another with genes outside that same pathway.
Discussion
We have built two models that perform similarly, which is shocking as we are trying to predict mutation in a specific gene. We would expect that the model with genes of the same pathway would perform better as it should add more information about genes that are not in the same pathway.
However, we found that the second one, with genes not in the pathway, does not perform worse but slightly better. So now we can ask ourselves, how does that make sense? Is there any information we do not know about that the neural network can see?. In this case, we could delve more into this topic and perform an extensive biological analysis, but what if we keep it simple?.
Our approach is knowing that we have two sets of genes that interact with KRAS in some manner; why don't we represent this interaction as a network?. We have used Cytoscape and Genemania addon to build a network using this two genes list:

In the network, the genes are represented by color depending on their status in the pathway. For example, in red are the genes inside the pathway, in blue those outside it, the genes in dark grey are the three central genes of the pathway, and in grey are additional genes added by Genemania to the network. Also, the circles represent significant subnetworks within the whole network.
The existing links between each node of the network represent if they are coexpressed and how significant this coexpression is. A pair of coexpressed genes implies that two genes have a similar pattern in their expression. Also, we have data about pathways inside our network, physical and genetic interaction, co-localization, predicted interactions, and shared protein domain. Ultimately, the most significant information from our data is the coexpression network we built.
Thus, with this analysis, we have found an explanation for how our models perform similarly. Furthermore, the neural network can detect this pattern and obtain information from it to build a successful model. So, in the end, we obtain two models able to predict if the pathway is altered, and therefore KRAS is mutated only from expression data.
Recommend
About Joyk
Aggregate valuable and interesting links.
Joyk means Joy of geeK